
OK, that’s a little bit over the top. I was trying to find a way to work NPH into this post. 🙂
Viruses are the simplest biological forms. Viruses are obligate parasites. A virus must invade a host cell in order to reproduce. Viruses contain genetic material, but do not have ribosomes which replicate the genetic material. Viruses use ribosomes in the host cell for replication.
Since a virus cannot reproduce on its own, one may ask if a virus is alive. The question “What is life?” is non-trivial and is not as easy to answer as you may think. Another interesting question to consider is “How did viruses and life begin?”
Please note the term “genetic material.” Viruses are exceptionally diverse. Some viruses are DNA-based, some are RNA-based. Further, the genetic material may be single-stranded or double-stranded. On top of these characteristics, the material may have a linear or a circular structure, or be segmented or non-segmented. Amazing.
RNA-based retroviruses are a huge exception to Francis Crick’s central dogma: DNA makes RNA and RNA makes protein. In a retrovirus, reverse transcriptase (a chemical enzyme/catalyst) synthesizes a DNA copy from viral RNA. The DNA copy integrates into the host genome (in eukaryotic cells). Then, the cell’s protein and replication factory takes over. Eventually, the cell bursts or otherwise expunges the particles and a new cells are infected.
Both ribonucleic acid (RNA) and deoxyribonucleic acid (DNA) are large molecules (polymers) — chains of nucleic acid molecules. Genetic material (RNA or DNA) is rather fragile. Strong UV light or heat causes the chemical bonds in RNA and DNA to break. Whether it’s a cell or a virus, genetic material needs to be protected. A virus typically has a hard protein “shell”, called a “capsid,” that surrounds and protects the genetic material inside.
At this point, I recommend the NOVA Labs and Khan Academy tutorial about RNA before reading on. They do quite a decent job of describing RNA, transcription (copying), and protein synthesis. If you want to cut to the chase, jump to the Virus Wars section of the tutorial.
SARS-CoV-2
SARS-CoV-2 is the virus which causes COVID-19, the disease. It is a type of coronavirus. Calling SARS-CoV-2 “coronavirus” is really refering to this specific type of virus by a generic name. (Not quite correct semantically.)
Coronaviruses contain positive-stranded RNA. The RNA is surrounded by an envelope with spike glycoproteins on the envelope. Without a doubt, you’ve seen the SARS-CoV-2 protein model on television — a grey envelope with red spikes. The spikey glycoproteins are composed of two subunits (S1 and S2). The spikes assist binding to a host cell and injection.
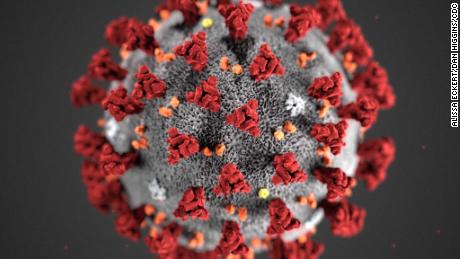
SARS-CoV-2 is approximately 60 to 140 nm in diameter. That’s darned small. A human hair has a diameter of 15 to 181 um, 1000 times larger. The bactrium E. coli is 1 to 2 um in length, still much larger than SARS-CoV-2. Viruses are exceptionally small; they must be small enough to invade host cells like E.coli.
SARS-CoV-2 has a single-stranded RNA genome. Its RNA strand contains 29891 nucleotides and encodes for 9860 amino acids. In biology-speak, that’s almost 30 kb (kilobases, not kilobytes). AS RNA-based viruses go, that’s a fairly large genome.
The folks at Fusion Medical Animation have produced a new SARS-CoV-2 animation. The image below is taken from their animation.
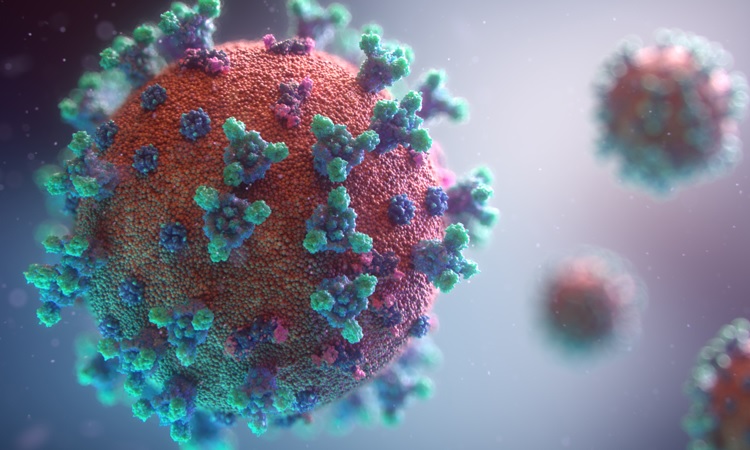
A tour of the genome
The New York Times published an excellent tour of the SARS-CoV-2 genome — Bad News Wrapped in Protein: Inside the Coronavirus Genome .
The Nucleocapsid gene (N), Envelope gene (E), Spike gene (S) and RNA-dependent RNA polymerase (RdRP) genes are important for testing. The WHO and CDC have developed nucleic acid amplification tests (NAAT) via reverse-transcription polymerase chain reaction (RT-PCR). Targeted genes include the N, E, S, and RdRP genes in the SARS-CoV-2 genome (WHO guidance). RT-PCR amplifies minute quantities of extracted SARS-CoV-2 RNA in a sample (if present) making it possible to assay one or more of the targeted subsequences (genes).
By now, you’ve come a long way along the journey and are ready for the summit. In closing, I recommend Features, Evaluation and Treatment Coronavirus (COVID-19) by Marco Cascella, et al. from the Istituto Nazionale Tumori – IRCCS – Fondazione Pascale. This paper is published by the National Center for Biotechnology Information (NCBI), a part of the U.S. National Institutes of Health (NIH).
If you would like to try your hand at sequencing, you can download the SARS-CoV-2 genome from NCBI. Don’t forget to snag a copy of BLAST. “BLAST finds regions of similarity between biological sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance.” STEM teachers, please take note of these resources. NCBI has many other genomes available for download.
Stay healthy, friends — P.J. Drongowski
